imager, an R package for image processing, has been updated to v0.20 on CRAN. It’s a major upgrade with a lot of new features, better documentation and a more consistent API.
imager now has 130 functions, and I myself keep forgetting all that’s in there. I’ve added a tutorial vignette that should help you get started. It goes through a few basic tasks like plotting and histogram equalisation and builds up to a multi-scale blob detector. It also covers plotting with ggplot2 and has a thematic list of functions.

New features added in the last months include new assignment functions, a utility for getting information on image files (iminfo), auto-thresholding based on k-means, much better array subset operators, updated docs and a reorganised codebase. Windows support should also have improved. Last but not least, you can now interrupt lengthy computations by hitting Ctrl+c or the stop button in RStudio.
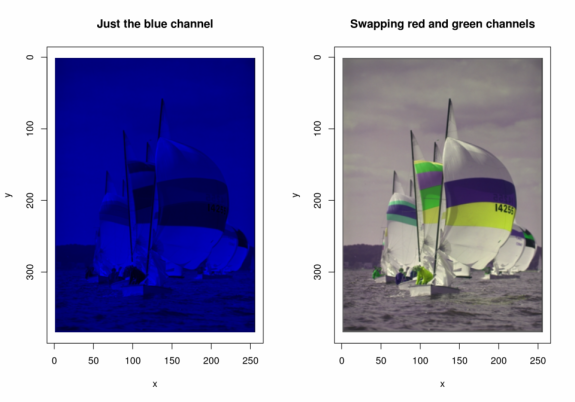
imager now has some easy-to-use replacement functions, meaning you can now do set image channels or change frames using a convenient R-like syntax:
library(imager)
boats.cp = boats #Make a copy of the boats image
R(boats.cp) = 0 #Set red channel to 0
G(boats.cp) = 0 #Set blue channel to 0
plot(boats.cp,main="Just the blue channel")
R(boats.cp) = G(boats)
G(boats.cp) = R(boats)
plot(boats.cp,main="Swapping red and green channels")
see ?imager.replace for more.
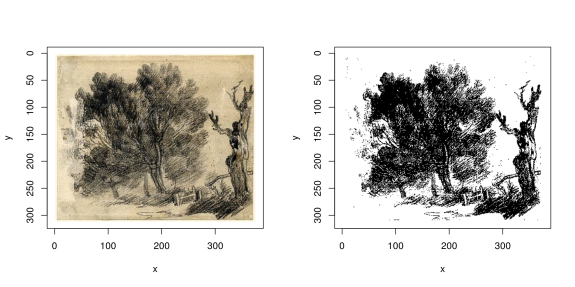
Auto-thresholding finds an optimal threshold for converting an image to binary values, based on k-means (it’s essentially a variant of Otsu’s method).
Here’s an illustration on a sketch by Thomas Gainsborough:
url = "https://upload.wikimedia.org/wikipedia/commons/thumb/3/30/Study_of_willows_by_Thomas_Gainsborough.jpg/375px-Study_of_willows_by_Thomas_Gainsborough.jpg"
im <- load.image(url)
layout(t(1:2))
plot(im)
grayscale(im) %>% threshold %>% plot
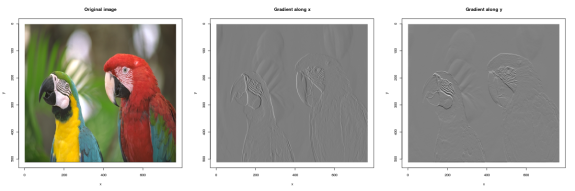
Point-wise reductions are useful for combining a list of images into a single output image. For example, enorm(list(A,B,C)) computes  , ie. the Euclidean norm. Here’s how you can use it to compute gradient magnitude:
, ie. the Euclidean norm. Here’s how you can use it to compute gradient magnitude:
imgradient(im,"xy") %>% enorm %>% plot("Gradient magnitude")

See also parmax, parmin, add, etc .
A note on compiling imager: if for some reason R tries to install imager from source (Linux or Mac), you will need the fftw library. On a Mac the easiest way is to grab it via Homebrew (“brew install fftw”), in Ubuntu “sudo apt-get install libfftw3-dev” should do it.